by Forrest Sheng Bao http://fsbao.net
I am so excited today that I have found a very cool Python library for doing wavelet decomposition, the PyWavelets. It is developed by Filip Wasilewski
To do 1D wavelet decomposition, just use the function :
wavedec(data, wavelet, mode='sym', level=None)where "data" is an array storing voltages of all sampling instant, "wavelet" is type of wavelet (e.g. Haar), "mode" is signal extension model(by default it's symmetric), "level" is the level of decomposition(by default it's 0).
The return value is an array of approximation (cA) and detail (cD) coefficients. These coefficients are just what I need to train the classifier.
Here is an example:
>>> import pywt
>>> coeffs = pywt.wavedec([1,2,3,4,5,6,7,8], 'db2', level=2)
>>> print pywt.waverec(coeffs, 'db2')
[ 1. 2. 3. 4. 5. 6. 7. 8.]
You can also do 2D wavelet decomposition by the function wavedec2. It's input are the same as 1D wavelet decomposition function wavedec This function will return a coefficients list [cAn, (cHn, cVn, cDn), ..., (cH1, cV1, cD1)], where n denotes the level of decomposition and cA, cH, cV and cD are approximation, horizontal detail, vertical detail and diagonal detail coefficients arrays. Here is an example:
>>> import pywt, numpy
>>> coeffs = pywt.wavedec2(numpy.ones((8,8)), 'db1', level=2)
>>> cA2, (cH2, cV2, cD2), (cH1, cV1, cD1) = coeffs
I also found another useful tool, pywfdb. It is used to read the data from the famous medical database, PhysioBank, physiologic signal archives for biomedical research. There are lots of open-access data for physiology, as well as neurology research. You can consider it as an Python implementation to WFDB, an open source package for viewing, analyzing, and creating recordings of physiologic signals. Flip gives an example of using his functions to read and plot a series of ECG data: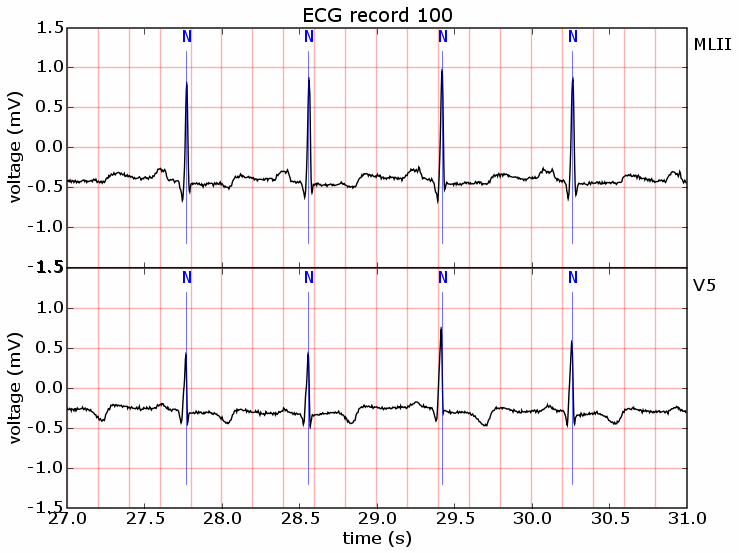
No comments:
Post a Comment